sdSuSiE Overview
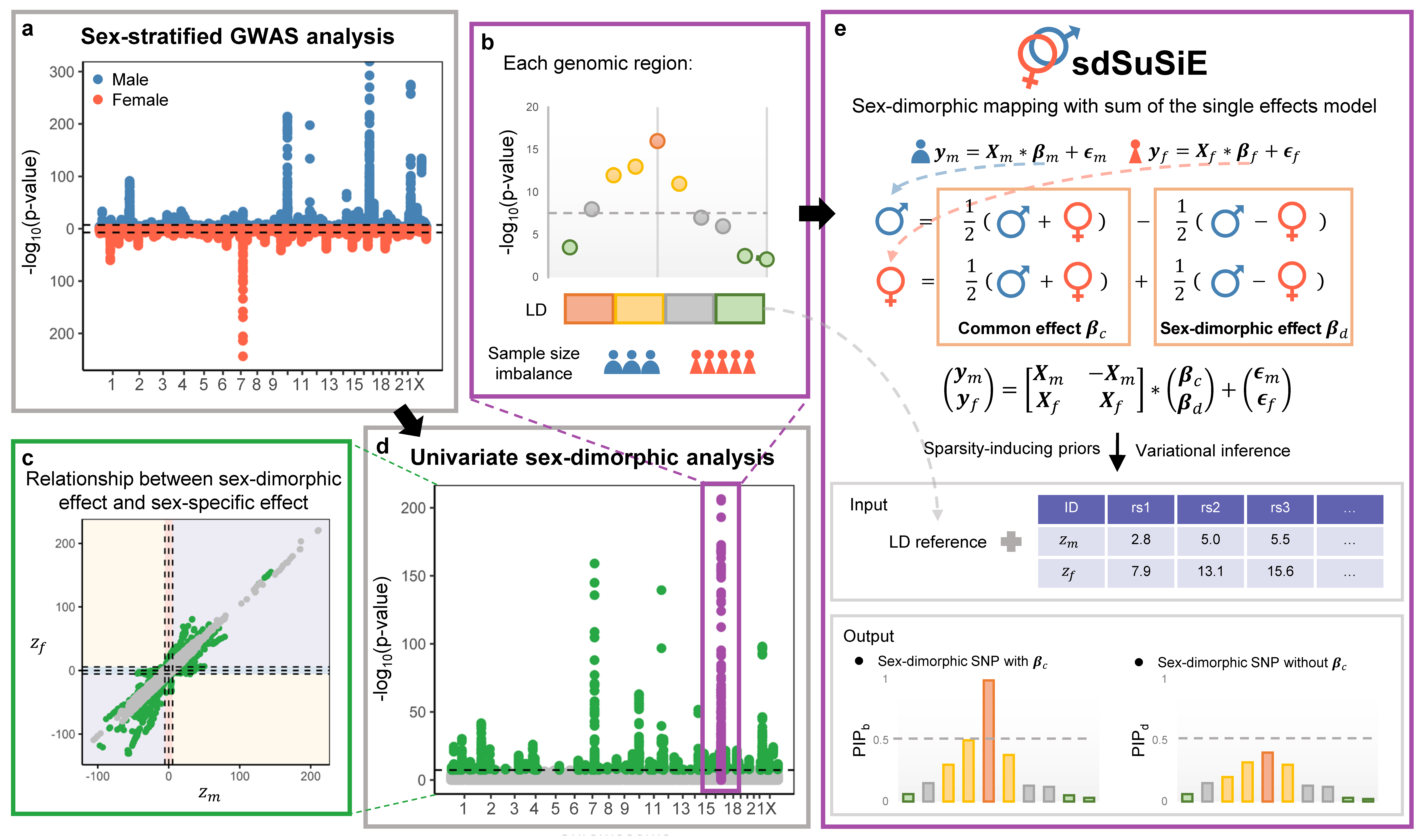 sdSuSiE is a package designed for sex-dimorphic fine-mapping using summary statistics from sex-stratified genome-wide association studies (GWAS). sdSuSiE explicitly models sex differences in genetic effects while accounting for correlations induced by linkage disequilibrium and sample size imbalances between sexes. It uses summary statistics from sex-stratified GWASs as inputs and extends the recent scalable variational inference algorithm SuSiE to fine-map sex-dimorphic effects. As a result, sdSuSiE provides calibrated credible set coverage and posterior inclusion probabilities (PIPs).
sdSuSiE is a package designed for sex-dimorphic fine-mapping using summary statistics from sex-stratified genome-wide association studies (GWAS). sdSuSiE explicitly models sex differences in genetic effects while accounting for correlations induced by linkage disequilibrium and sample size imbalances between sexes. It uses summary statistics from sex-stratified GWASs as inputs and extends the recent scalable variational inference algorithm SuSiE to fine-map sex-dimorphic effects. As a result, sdSuSiE provides calibrated credible set coverage and posterior inclusion probabilities (PIPs).
Installation
You can install the released version of sdSuSiE from Github with the following code, for more installation details or solutions that might solve related issues (specifically MacOS system) see the link.
Dependencies
- R version >= 3.5.0.
- R packages: R6, nloptr, cowplot, dplyr, ggplot2, ggrepel, Rcpp, RcppArmadillo, parallel, doParallel, foreach
1. Install devtools if necessary
install.packages('devtools')
2. Install sdSuSiE
devtools::install_github('yuanzhongshang/sdSuSiE')
3. Load package
library(sdSuSiE)
This package is supported for Windows 10, MAC and Linux. The package has been tested on the following systems:
- Windows 10
- MAC: OSX (10.14.1)
- Linux: Ubuntu (16.04.6)
Issues
All feedback, bug reports and suggestions are warmly welcomed! Feel free to leave messages on the github issues. Please make sure to raise issues with a detailed and reproducible example and also please provide the output of your sessionInfo() in R!
How to cite sdSuSiE
Lu Liu, Zhongshang Yuan, Xiang Zhou, Scalable and powerful sex-dimorphic fine-mapping with summary statistics from sex-stratified genome-wide association studies, 2025.
How to use sdSuSiE
A quick guide with an example analysis: here.