GIFT Overview
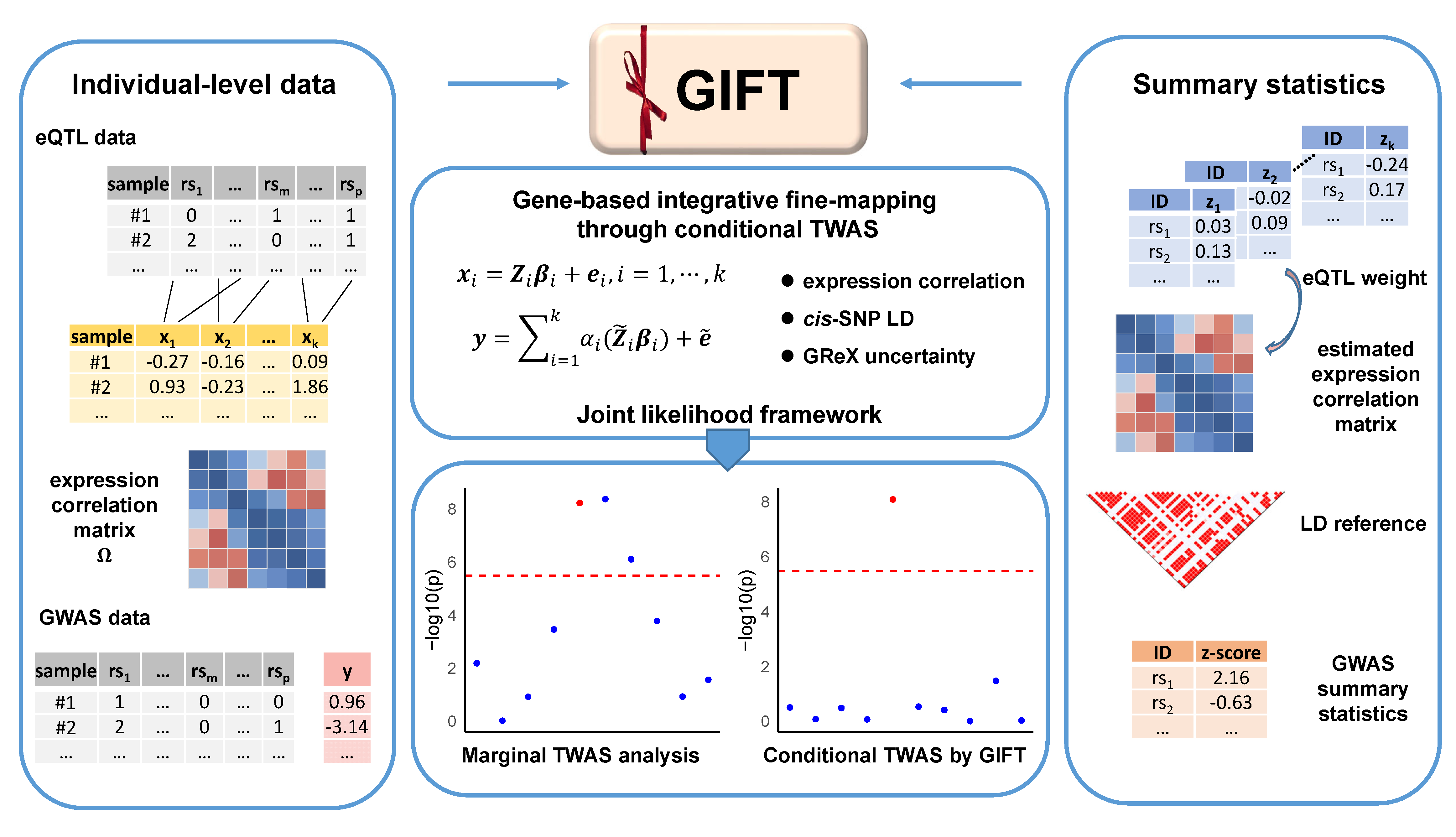 GIFT is an R package for efficient statistical inference of conditional TWAS fine-mapping. GIFT examines one genomic region at a time, jointly models the GReX of all genes residing in the focal region, and carries out TWAS conditional analysis in a maximum likelihood framework. In the process, GIFT explicitly models the gene expression correlation and cis-SNP LD across different genes in the region, accounts for the uncertainty in the constructed GReX through joint inference and provides calibrated p values. GIFT is implemented as an open-source R package, freely available at www.xzlab.org/software.html.
GIFT is an R package for efficient statistical inference of conditional TWAS fine-mapping. GIFT examines one genomic region at a time, jointly models the GReX of all genes residing in the focal region, and carries out TWAS conditional analysis in a maximum likelihood framework. In the process, GIFT explicitly models the gene expression correlation and cis-SNP LD across different genes in the region, accounts for the uncertainty in the constructed GReX through joint inference and provides calibrated p values. GIFT is implemented as an open-source R package, freely available at www.xzlab.org/software.html.
Installation
You can install the released version of GIFT from Github with the following code, for more installation details or solutions that might solve related issues (specifically MacOS system) see the link.
Dependencies
- R version >= 4.0.0.
- R packages: Rcpp, RcppArmadillo, parallel
1. Install devtools if necessary
install.packages('devtools')
2. Install GIFT
devtools::install_github('yuanzhongshang/GIFT')
3. Load package
library(GIFT)
This package is supported for Windows 10, MAC and Linux. The package has been tested on the following systems:
- Windows 10
- MAC: OSX (10.14.1)
- Linux: Ubuntu (16.04.6)
Issues
All feedback, bug reports and suggestions are warmly welcomed! Please make sure to raise issues with a detailed and reproducible example and also please provide the output of your sessionInfo() in R!
How to cite GIFT
Liu, L., Yan, R., Guo, P. et al. Conditional transcriptome-wide association study for fine-mapping candidate causal genes. Nat Genet 56, 348–356 (2024). https://doi.org/10.1038/s41588-023-01645-y
How to use GIFT
Example Analysis with GIFT: here.
The GIFT Manual: here.
The genome-wide eQTL summary statistics from GEUVADIS data
This data is availible in the dropbox: here.
The correlation matrix among gene expressions for each chromosome from GEUVADIS data is also availible in the dropbox: here.